Introduction
The well-known double helix carries the DNA of living beings. The human DNA contains about 3.2 billion nucleotide base pairs represented by the quaternary symbols (A, G, C, T). With high-speed sequencing machines today it is possible to “read” the DNA. The resulting file contains millions of “reads”, short segments of symbols, typically all of the same length, and weighs an unwieldy few Terabytes.
The upcoming MPEG-G standards, developed jointly by MPEG and ISO TC 276 Biotechnology, will reduce the size of the file, without loss of information, by exploiting the inherent redundancy of the reads and make at the same time the information in the file more easily accessible.
This article provides some context, and explains the basic ideas of the standard and the benefits it can yield to those who need to access genomic information.
Reading the DNA
There are two main obstacles preventing a direct use of files from sequencing machines: the position of a read on the DNA sample is unknown and the value of each symbol of the read is not entirely reliable.
The picture below represents a 17 reads with a read length of 15 nucleotides. These have been aligned to a reference genome (first line). Reads with a higher number start further down in the reference genome.

Reading column-wise, we see that in most cases the values have exactly the value of the reference genome. A single difference (represented by isolated red symbols) may be caused by read errors while a quasi completely different column (most symbols in red) may be caused by the fact that 1) a given DNA is unlikely to be exactly equal to a reference genome or 2) the person with this particular DNA may have health problems.
Use of genomics today
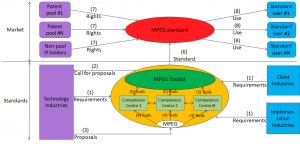
Genomics is already used in the clinical practice. An example of genomic workflow is depicted in the figure below which could very well represent a blood test workflow if “DNA” were replaced by “blood”. Patients go to a hospital where a sample of their DNA is taken and read by a sequencing machine. The files are analysed by experts who produce reports which are read and analysed by doctors who decide actions.
Use of genomics tomorrow
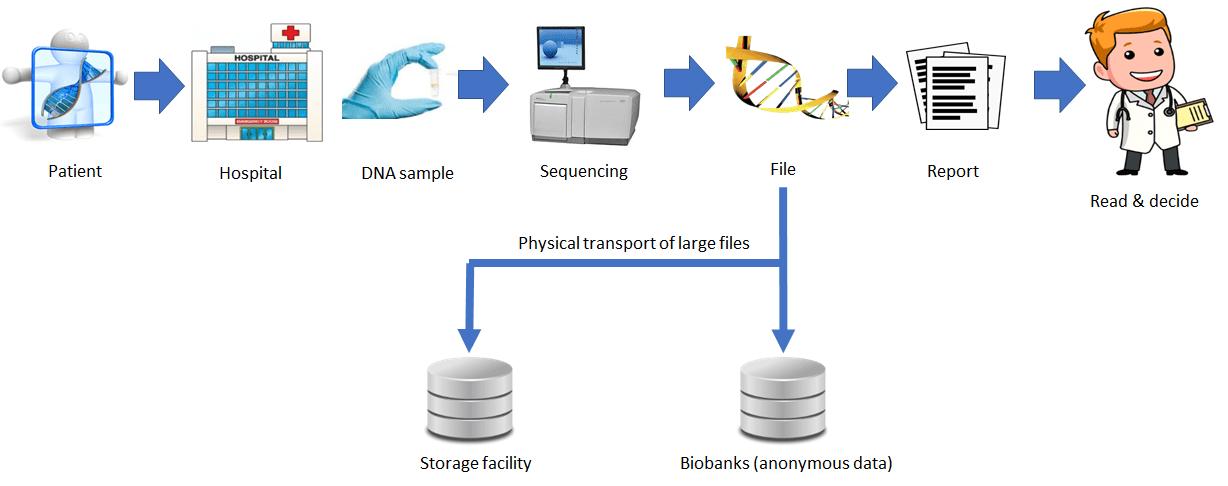
Today genomic workflows take time – even months – and are costly – thousands of USD per DNA sample. While there is not much room to cut the time it takes to obtain a DNA sample, sequencing cost has been decreasing and are expected to continue doing so.
Big savings could be achieved by acting on data transport and processing. If the size of a 3 Terabytes file is reduced by, say, a factor of 100, the transport of the resulting 30 Gigabytes would be compatible with today’s internet access speeds of 1 Gbit/s (~4 min). Faster data access, a by-product of compression, would allow doctors to get the information they are searching, locally or from remote, in a fraction of a second.
The new possible scenario is depicted in the figure below.
MPEG makes genome compression real
Not much had been done to make the scenario above real (zip is the oft-used compression technology today) until the time (April 2013) MPEG received a proposal to develop a standard to losslessly compress files from DNA sequencing machines.
The MPEG-G standard – titled Genomic Information Representation – has 5-parts: Parts 1 and 2 are expected to be approved at MPEG 125 (January 2018) and the other parts are expected to follow suit shortly after.
MPEG-G is an excellent example of how MPEG could apply its expertise to a different field than media. Part 1, an adaptation of the MP4 File Format present in all smartphones/tablets/PCs, specifies how to make and transport compressed files. Part 2 specifies how to compress reads and Part 3 how to invoke the APIs to access specific compressed portions of a file. Part 4 and 5 are Conformance and Reference Software, respectively.
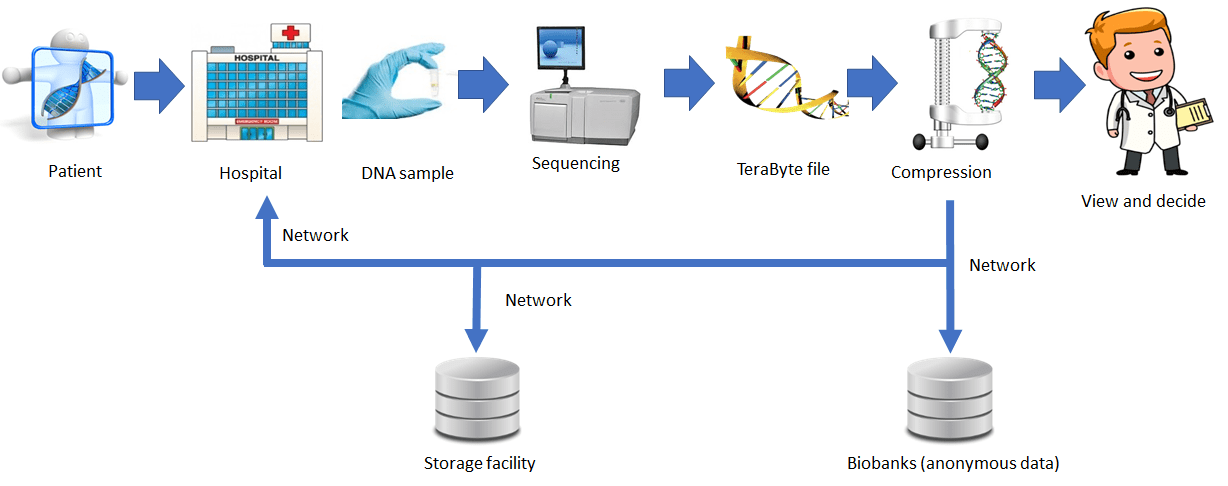
The figure below depicts the very sophisticated operation specified in Part 2 in a simplified way.
An MPEG-G file can be created with the following sequence of operations:
- Put the reads in the input file (aligned or unaligned) in bins corresponding to segments of the reference genome
- Classify the reads in each bin in 6 classes: P (perfect match with the reference genome), M (reads with variants), etc.
- Convert the reads of each bin to a subset of 18 descriptors specific of the class: e.g., a class P descriptor is the start position of the read etc.
- Put the descriptors in the columns of a matrix
- Compress each descriptor column (MPEG-G uses the very efficient CABAC compressor already present in several video coding standards)
- Put compressed descriptors of a class of a bin in an Access Unit (AU) for a maximum of 6 AUs per bin
Therefore MPEG-G file contains all AUs of all bins corresponding to all segments of the reference genome. A file may contain the compressed reads of more than one DNA sample.
The benefits of MPEG-G
Compression is beneficial but is not necessarily the only or primary benefit. More important is the fact that while designing compression, MPEG has given a structure to the information. In MPEG-G the structure is provided by Part 1 (File and transport) and by Part 2 (Compression).
In MPEG-G most information relevant to applications is immediately accessible, locally and, more importantly, also from remote without the need to download the entire file to be able to access the information of interest. Part 3 (Application Programming Interfaces) makes this fast access even more convenient because it facilitates the work of developers of genomics applications who may not have in-depth information of the – certainly complex – MPEG-G standard.
Conclusions
In the best MPEG tradition, MPEG-G is a generic standard, i.e. a standard that can be employed in a wide variety of applications that require small footprint of and fast access to genomic information.
A certainly incomplete list includes: Assistance to medical doctors’ decisions; Lifetime Genetic Testing; Personal DNA mapping on demand; Personal design of pharmaceuticals; Analysis of immune repertoire; Characterisation of micro-organisms living in the human host; Mapping of micro-organisms in the environment (e.g. biodiversity).
Standards are living beings, but MPEG standards have a DNA that allows them to grow and evolve to cope with the manifold needs of its ever-growing number of users.
I look forward to welcoming new communities in the big family of MPEG users.
Posts in this thread (in bold this post)
- The MPEG ecosystem
- Why is MPEG successful?
- MPEG can also be green
- The life of an MPEG standard
- Genome is digital, and can be compressed
- Compression standards and quality go hand in hand
- Digging deeper in the MPEG work
- MPEG communicates
- How does MPEG actually work?
- Life inside MPEG
- Data Compression Technologies – A FAQ
- It worked twice and will work again
- Compression standards for the data industries
- 30 years of MPEG, and counting?
- The MPEG machine is ready to start (again)
- IP counting or revenue counting?
- Business model based ISO/IEC standards
- Can MPEG overcome its Video “crisis”?
- A crisis, the causes and a solution
- Compression – the technology for the digital age
- On my Charles F. Jenkins Lifetime Achievement Award
- Standards for the present and the future